-
Chromosome evolution in grasshoppers
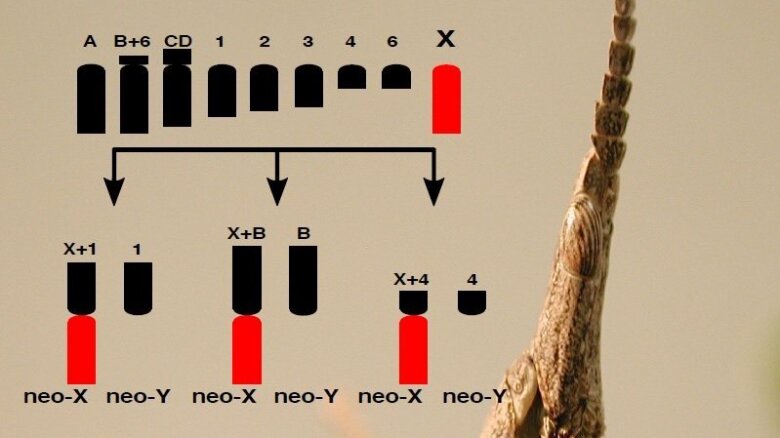
Sex is a fundamental and ancient feature of eukaryotic reproduction often associated with the presence of specialized sex chromosomes involved in female or male development. Despite the importance and conservation of sexual reproduction, there is a notable diversity of sex chromosomes within and between sexes: XY system with female XX and male XY, and ZW system with female ZW and male ZZ. This diversity is likely to have key consequences for multiple facets of evolution, as sex chromosomes play central roles in adaptation, speciation and sexual dimorphism but remains unclear how sex chromosomes are built and what kind of sex-specific changes occur. Understanding the causes and consequences of sex chromosome evolution requires study systems where sex chromosomes have evolved recently and independently several times. My current research involve the understanding of the early signatures of sex chromosome evolution using the grasshopper Vandiemenella viatica species complex. While most species have and X0 system, where females have two X chromosomes and males only one, there has been repeated and independent chromosomal fusions between the ancestral X and autosomes resulting in the formation of new Y chromosomes. By combining state-of-the art genomics, transcriptomics, single cell resolution, and cytogenetics, we seek to provide an integrated understanding of the early signatures of sex chromosome evolution. The research aims to answer three key questions:
- How is recombination suppressed between nascent sex chromosomes?
- How does gene regulation evolve after recombination suppression?
- How fast does the neo-Y degenerate after recombination suppression?
Funding
Starting Grant of the Swedish Research Council, 2021-2023; Grant of the Carl Trygger Foundation, 2022-2024.People involved
Octavio M. Placios-Gimenez, Suvratha JayaprasadNeo-sex chromosome system in Vandiemenella
Image: Octavio M. Palacios-Gimenez -
The evolutionary ecology of individualized niches
Learn moreExternal linkGrasshopper Diversity
Image: Holger SchielzethIndividuals differ. This seemingly trivial statement has led to paradigm shifts, as three different fields of organismal biology have seen a marked change in key concepts over the last years. In behavioral biology, it has been realized that there are profound differences between individuals and that these can be stable over time and across contexts, giving rise to the concept of animal personalities. In ecology, an increasing focus is likewise on the considerable variation in the ecological niche realized by species, populations, and even individuals. In evolutionary biology, where individual variation has always been central, there is an increasing awareness of the complexity with which genotypes interact with the environment to produce unique phenotypes. As a consequence, a concept of an individualized niche is needed, rather than focusing only on a mean value for a given population.
The central research goal of the collaborative research center is to redefine the niche concept on the individual level. By doing so, we want to gain a comprehensive understanding of how individual phenotypes interact with their environment and what the ensuing consequences for ecological and evolutionary processes are. We hypothesize that, across taxa, the interaction between the individualized phenotype and the environment results in individualized niches via three mechanisms of adjustment and adaptation: niche choice, niche conformance and niche construction.
Funding
Collaborative research center SFB Transregio 212 funded by the German Research Foundation (DFG)External link, 2018-2021People involved
Holger Schielzeth, Gabe Winter, Elina Takola, Martin StoffelPublications
Moran, N., Sánchez-Tójar, A., Schielzeth, H. & Reinhold, K. (2021). Poor nutritional condition promotes high-risk behaviours: A systematic review and meta-analysis. Biological Reviews 96: 269-288. doi: 10.1111/brv.12655External link
Köhler, G. & Schielzeth, H. (2020). Green-brown polymorphism in alpine grasshoppers affects body temperature. Ecology and Evolution 10: 441-450. doi: 10.1002/ece3.5908External link
Schmoll, T., Rudolfsen, G., Schielzeth, H. & Kleven, O. (2020). Sperm velocity in a promiscuous bird across experimental media of different viscosities. Proceedings of the Royal Society B 287: 20201031. doi: 10.1098/rspb.2020.1031External link
Voelkl, B., Altman, N.S., Forsman, A., Forstmeier, W., Gurevitch, J., Jaric, I., Karp, N.A., Kas, M.J., Schielzeth, H., Van de Casteele, T. & Würbel, H. (2020). Reproducibility of animal research in light of biological variation. Nature Reviews Neuroscience 21: 384-393. doi: 10.1038/s41583-020-0313-3External link
-
Molecular basis of phenotypic variation in wheatears
Red-tailed Wheatear (Oenanthe chrysopygia)
Image: Reto BurriWheatears (genus Oenanthe) are an about 6.6 my old group of passerine birds mainly inhabiting arid and rocky ecosystems of Eurasia and Africa that exhibit striking patterns of phenotypic polymorphism: (i) High rates of character switching across the phylogeny suggests multiple convergent origins of numerous phenotypes, including melanin-based coloration and complex behavioral traits such as seasonal migration. (ii) Multiple interspecific differences segregate as polymorphisms within species. And finally, (iii) one of these polymorphisms found in black-eared wheatear (O. hispanica) and pied wheatear (O. pleschanka) supposedly arose by reciprocal introgression among these two pervasively hybridizing sister species. The convergent emergence of phenotypes across the phylogeny both within and between species points towards an involvement of a labile molecular switch between phenotypes.
We aim at addressing multiple questions relating to the evolution of phenotypic diversity within and between species, and to the evolution of species. Currently we settled out to characterize the genetic population structure across the hybrid zone of black-eared and pied wheatear using genome-wide polymorphism data. In future phenotypic, genetic, and methylomic polymorphism data from within these species, from three hybrid zones, and from across the wheatear genus will be used to identify the molecular bases of diverse color phenotypes and study their evolution, including the demographic and genomic constrains under which they evolve.
Funding
Research Grant BU-3456/3-1External link by the German Research Foundation (DFG), 2018-2021
Postdoctoral fellowship of the Alexander von Humboldt FoundationExternal link to Niloo Alaei Kakhki, 2019-2021People involved
Niloofar Alaei Kakhki, Reto Burri, Dave LutgenPublications
Lutgen, D., Ritter, R., Olsem, R.-A., Schielzeth, H., Gruselius, J., Ewels, P., García, J.T., Shirihai, H., Schweizer, M., Suh, A. & Burri, R. (2020). Linked-read sequencing enables haplotype-resolved resequencing at population scale. Molecular Ecology Resources 20: 1311-1322. http://dx.doi.org/10.1111/1755-0998.13192
Schweizer, M., Warmuth, V., Alaei Kakhki, N., Aliabadian, M., Förschler, M., Shirihai, H., Suh, A. & Burri, R. (2019). Parallel plumage colour evolution and introgressive hybridization in wheatears. Journal of Evolutionary Biology 31: 100-110. http://dx.doi.org/10.1111/jeb.13401
Schweizer, M., Warmuth, V.M., Alaei Kakhki, N., Aliabadian, M., Förschler, M., Shirihai, H., Ewels, P., Gruselius, J., Olsen, R.A., Schielzeth, H., Suh, A. & Burri, R. (2019). Genome-wide evidence supports mitochondrial relationships and pervasive parallel phenotypic evolution in open-habitat chats. Molecular Phylogenetics and Evolution 139: 106568. http://dx.doi.org/10.1016/j.ympev.2019.106568
-
Origins of divergence in Spodoptera strains
Speciation is the evolutionary process by which new biological species arise. There has been increasing acceptance of sympatric speciation models, where reproductive isolation and incompatibility between populations evolve in the face of gene flow; yet documenting this process remains challenging since gene flow reduction most likely results from a combination of factors. The reproductive barriers can be initiated either by divergent selection (i.e., ‘ecological’ or sexual selection) or by the evolution of genetic incompatibilities, i.e. through genetic drift, as an indirect consequence of selection, or through genomic conflict.This project aims at deciphering the factors at play in the divergence of the two sympatric strains of the fall armyworm (FAW, Spodoptera frugiperda). This noctuid moth is an ideal model organism to investigate different isolation barriers in incipient species. While morphologically indistinguishable, the two strains can be differentiated on the basis of genetic haplotypes, and they exhibit a number of phenotypic differences that can function as pre-or post-zygotic isolation barriers. The objectives of the project are:1) Identification and characterization of diverging genetic loci between the strains.Using a new population genomics approach, all loci diverging between the strains will be identified. The variants will be classified according to the hypothetical contribution to speciation: timing, oviposition preference, pheromones, female hybrid frigidity larval preference, larval performance, female hybrid fertility. They will be functionally characterized using CRISPR/CAS9.2) Determination of the origin of post-zygotic reproductive isolation in invasive populations.Preliminary data of invasive populations in Africa suggests fitness costs for corn-collected larvae on rice and vice versa, illustrating FAWs adaptive potential in invaded areas. We will analyze the genomic differentiation of these invasive populations.3) To investigate the causes of strain-specific host plant differentiationThe differential distribution of the strains on different host plants in native habitats likely result from different oviposition choices or from differences in egg quality. We will determine the genetic basis of these traits by GWAS.4) To determine the ultimate causes of the strain-specific timing differentiationThe strains differ by their mating time at night. We will test experimentally if these differences can be related to variation in biotic factors connected to different habitats, e.g parasitoids or predator occurrence or plant defense rhythms.This unique integrative study is based on the combination of various approaches: The study of phenotypes in natura and forward genetics in Germany and reverse genetics and functional genomics in France. It will clarify evolutionary status of the FAW strains and is thus a prerequisite to understand the success of the invasive populations in their new environment.
Funding
Research GrantExternal link by German Research Foundation (DFG)External link, 2020-2024.People involved
Sabine Hänniger -
Reticulated speciation in Chorthippus grasshoppers
The ‘omics’ era reveals an increasing number of species that are practically indistinguishable in morphology, ecology or even in genetics, despite being reproductively isolated from one another in sympatry. While such species complexes present an example of taxonomic conundrum for systematists, they are prized as valuable study systems for evolutionary biologists because the behavioral traits that maintain species boundaries can directly be observed. Here, we propose to study one of such systems, sympatric grasshopper species of the genus Chorthippus that have radiated under pervasive gene flow. This offers opportunities for identifying the genes underlying behavioral isolation and speciation. Using a comparative phylogeographic approach we will test whether species have diverged with ongoing gene flow, or if geographic isolation during glacial periods has facilitated reproductive isolation. Using experimental crosses, we will test for a genetic association between cues and preference of behavioral isolation, which facilitates the maintenance of species boundaries in the face of gene flow. Lastly, using population genomics and behavioral assays, we will test if gene flow has played a role in generating hybrid species that are reproductively isolated from their parentals. This research program will not only offer new insights on how gene flow interacts with sexual selection during species formation, but it will provide a transferable multi-disciplinary approach for taxonomic studies in organisms characterized by large genomes that remain challenging to study.
Funding
Research GrantExternal link by German Research Foundation (DFG)External link, 2022-2024.People involved
Ricardo PereiraExternal link, Dörte NeumeisterExternal link, Holger Schielzeth -
The Jena Experiment
Learn moreExternal linkJena Experiment Weeding
Image: Gerlinde KratzschThe Jena ExperimentExternal link is one of the longest-running biodiversity experiments worldwide. It was designed to study the functional consequences of grassland biodiversity. Experimental plots were established already in 2002 from a pool of 60 local grassland plant species assembled in experimental communities ranging from monocultures to 60-species mixtures. The field site in the Saale floodplain currently hosts more than 300 experimental plots in an area of 10 ha. A videoExternal link provides an excellent overview.
The Jena Experiment has provided important insights into the links between biodiversity, population processes in grassland communities, and ecosystem fluxes and services. Specifically, it has demonstrated positive effects of plant diversity on plant productivity and a large number of other ecosystem variables, supporting the hypothesis that biodiversity maintains ecosystem multifunctionality. The value of the Jena Experiment has increased over time as now long-term plots allow capturing representative biodiversity effects and the unprecedented wealth of data enables unique syntheses and meta-analyses. An extensive paper published in Basic and Applied EcologyExternal link gives a comprehensive overview of the findings from the first 15 years of the experiment.
The Jena Experiment has always been a highly collaborative project that involves several national and international collaboration partners. It has been immensely productive in terms of scientific output with over 300 peer-reviewed scientific publications. Anne Ebeling from the Population Ecology Group has served as the scientific coordinator of the project since 2009.
In 2019, The German Research Foundation (DFG)External link has decided to fund a new Research Unit (FOR 5000External link) to address new questions in the Jena-Experiment. Specifically, the research unit aims to understand the biotic interactions, community assembly processes and eco-evolutionary dynamics that are potential drivers of long-term biodiversity–ecosystem functioning relationships. The research unit encompasses 11 scientific subprojects. Anne is involved as a principle investigator in the central coordination project and in a subproject on Plant-consumer interactions as cause and consequence of long-term BEF relationshipsExternal link. Holger is a principle investigator in a subproject on The effects of interspecific biodiversity on intraspecific genetic diversity: a community genomics approachExternal link.
Funding
Research Unit FOR 5000External link funded by the German Research Foundation (DFG)External link, 2020-2023. Previous funding: Research Unit FOR 1451External link (2010-2018) and Research Unit FOR 456External link (2002-2008) funded by the German Research Foundation (DFG)External linkPeople involved
Maximilian Bröcher, Anne Ebeling, Yuanyuan Huang, Annette Jesch, Holger Schielzeth, Chongyi Jiang -
Nutrient Network
Learn moreExternal linkNutNet Field Site
Image: Ilka WolfNutrient Network (NUTNET)External link One the most important impacts of human activity on grassland ecosystems is the application of fertilizers and the alteration of grazers-plant interactions. To study the effect of these activities, the Nutrient Network (NUTNET), a grassroots research effort, was established in 2007. This research network, coordinated by the Department of Ecology, Evolution, and Behaviour at the University of Minnesota, comprises more than 130 grassland sites worldwide, allowing global analyses focussing on the impact of fertilization and alteration of plant-consumer interactions on grassland ecosystems. The Jena NUTNET site, one of three in Germany, is coordinated by Anne Ebeling from the Population Ecology Group and Christiane Roscher from iDiv in Leipzig. In Jena we are especially interested in the consequences of fertilizer application for plant-consumer interactions and related ecosystem processes (e.g. leaf damage by arthropods).
People involved
Anne Ebeling -
Bug Network
Learn moreExternal linkWhile a few studies have shown big impacts of invertebrates and fungal pathogens on plant communities and ecosystems, we know little about how generally important they are, and how their functional composition varies across a range of different ecosystems.
As a globally coordinated research initiative the Bug Network is a powerful tool to investigate the context dependence of biotic interactions in grassland ecosystems. The use of standardized measurements and replicated experiments across ecological gradients allows direct comparison across sites. This is particularly important if we are to understand how global change drivers, such as climate and land use change, will alter consumer communities and their functioning.
In our group we contribute to the comparative part of the network (a cross-site survey). In two nearby grasslands we characterize plant communities, sample invertebrate communities and assess leaf damage caused by fungal pathogens and herbivores. Currently, as part of a master's thesis, we are particularly interested in the composition of beetle communities under different abiotic and biotic conditions.
For more details on the Bug Network, please see https://bug-net.org/External link.
People involved
Anne Ebeling -
Statistical quantification of individual differences
Statistical Quantification of Individual Differences is the product of the SQuID working group. The package aims to help scholars who, like us, are interested in understanding patterns of phenotypic variance. Individual differences are the raw material for natural selection to act on and hence the basis of evolutionary adaptation. Understanding the sources of phenotypic variance is thus a most essential feature of biological investigation. Mixed effects models offer a great, albeit challenging tool in this context. Disseminating the properties, potentials and interpretational challenges in the research community is thus a foremost goal of SQuID.
The squid package has two main objectives: First, it provides an educational tool useful for students, teachers and researchers who want to learn to use mixed-effects models. Users can experience how the mixed-effects model framework can be used to understand biological phenomena by interactively exploring simulated multilevel data. Second, squid offers research opportunities to those who are already familiar with mixed-effects models, as it enables the generation of datasets that users may download and use for a range of simulation-based statistical analyses such as power and sensitivity analysis of multilevel and multivariate data.
Funding
INPART program of the Norwegian Research Council, 2021-2023People involved
Holger Schielzeth and many international collaboratorsPublications
Schielzeth, H., Dingemanse, N., Nakagawa, S., Westneat, D.F., Allegue, H., Teplisky, C., Réale, D., Dochtermann, N.A., Garamszegi, L.Z. & Araya-Ajoy, Y.G. (2020). Robustness of linear mixed-effects models to violations of distributional assumptions. Methods in Ecology and Evolution 11: 1141-1152. doi: 10.1111/2041-210X.13434External link
Westneat, D.F., Araya-Ajoy, Y.G., Allegue, H., Class, B., Dingemanse, N., Dochtermann, N.A., Garamszegi, L.Z., Martin, J.G.A., Nakagawa, S., Réale, D. & Schielzeth, H. (2020). Collision between biological process and statistical analysis revealed by mean centring. Journal of Animal Ecology 89: 2813-2824. doi: 10.1111/1365-2656.13360External link
Allegue, H., Araya-Ajoy, Y.G., Dingemanse, N.J., Dochtermann, N.A., Garamszegi, L.Z., Nakagawa, S., Réale, D., Schielzeth, H. & Westneat, D.F. (2017). Statistical Quantification of Individual Differences (SQuID): an educational and statistical tool for understanding multilevel phenotypic data in linear mixed models. Methods in Ecology and Evolution 8: 257-267. doi: 10.1111/2041-210X.12659External link
Ein Video ueber das Jena-Experiment aus Anlass unseres Beitrags zum MINT-Festival 2021.
-
Genetics of sexual selection in grasshoppers
Large Gold Grasshopper (Chrysochraon dispar)
Image: Holger SchielzethAdaptive evolutionary change occurs when selection is acting on heritable trait variation. But not all evolutionary responses are straightforward. Genetic covariation in particular may modify the speed and the direction of adaptive evolution. Genetic covariation arises from pleiotropy (the same genetic factors influence multiple traits) or from linkage disequilibrium (coinheritance) of multiple independent genetic factors. This can affect multiple traits of the same individual, but also traits expressed in different individuals, such as traits expressed in females and males. We have therefore studied the multivariate genetic architecture of trait variation in multiple species of grasshoppers to evaluated if evolution in grasshoppers is constraint or shaped by genetic covariation.
Sexual selection is a particularly potent force that can result in the evolution of extravagant ornaments and is therefore a driving force in generating biological diversity. We have therefore focused our research on the highly sexually dimorphic club-legged grasshopper. The species is unusual in that males possess swollen front legs (‘Popeye arms’). Neither females nor any related species show this feature and this begs the question about how these structures are used and how they have evolved. We have therefore studied the behavioral ecology of sexual selection in this intriguing species. It turns out that the courtship behavior of this species is highly peculiar.
Funding
Independent Emmy Noether research groupExternal link funded by the German Research Foundation (DFG)External link, 2012-2017People involved
Anasuya Chakrabarty, Petra Dieker, Holger Schielzeth, Abhijeet Shah, Pablo Valverde (completed)Publications
Chakrabarty, A. & Schielzeth, H. (2020). Comparative analysis of the multivariate genetic architecture of morphological traits in three species of gomphocerine grasshoppers. Heredity 124: 367-382. doi: 10.1038/s41437-019-0276-1External link
Chakrabarty, A., van Kronenberg, P., Toliopoulos, N. & Schielzeth, H. (2019). Direct and indirect genetic effects on reproductive investment in a grasshopper. Journal of Evolutionary Biology 32: 331-342. doi: 10.1111/jeb.13417External link
Dieker, P., Beckmann, L., Teckentrup, J. & Schielzeth, H. (2018). Spatial analyses of two colour polymorphisms in an alpine grasshopper reveal a role of small-scale heterogeneity. Ecology and Evolution 8: 7273-7284. doi: 10.1002/ece3.4156External link
Köhler, G., Samietz, J. & Schielzeth, H. (2017). Morphological and colour morph clines along an altitudinal gradient in the meadow grasshopper Pseudochorthippus parallelus. PLoS One 12: e0189815. doi: 10.1371/journal.pone.0189815External link
Schielzeth, H. & Dieker, P. (2020). The green-brown polymorphism of the club-legged grasshopper Gomphocerus sibiricus is heritable and appears genetically simple. BMC Evolutionary Biology 20: 63. doi: 10.1186/s12862-020-01630-7External link
Schielzeth, H. & Husby, A. (2014). Challenges and prospects in genome-wide quantitative trait loci mapping of standing genetic variation in natural populations. Annals of the New York Academy of Sciences 1320: 35-57. doi: 10.1111/nyas.12397External link
Schielzeth, H., Rios Villamil, A. & Burri, R. (2018). Success and failure in replication of genotype-phenotype associations: How does replication help in understanding the genetic basis of phenotypic variation in outbred populations? Molecular Ecology Resources 4: 739-754. doi: 10.1111/1755-0998.12780External link
Schielzeth, H., Streitner, C., Lampe, U., Franzke, A. & Reinhold, K. (2014). Genome size variation affects song attractiveness in grasshoppers: evidence for sexual selection against large genomes. Evolution 68: 3629-3635. doi: 10.1111/evo.12522External link
Shah, A., Hoffman, J.I. & Schielzeth, H. (2020). Comparative analysis of genomic repeat content in gomphocerine grasshoppers reveals expansion of satellite DNA and helitrons in species with unusually large genomes. Genome Biology and Evolution 12: 1180-1193. doi: 10.1093/gbe/evaa119External link
Shah, A., Hoffman, J.I. & Schielzeth, H. (2019). Transcriptome assembly for a colour-polymorphic grasshopper (Gomphocerus sibiricus) with a very large genome size. BMC Genomics 20: 370. doi: 10.1186/s12864-019-5756-4External link
Shah, A.B., Schielzeth, H., Albersmeier, A., Kalinowski, J. & Hoffman, J.I. (2016). High throughput sequencing and graph-based cluster analysis facilitate microsatellite development from a highly complex genome. Ecology and Evolution 6: 5718-5727. doi: 10.1002/ece3.2305External link
Valverde, J.P., Eggert, H., Kurtz, J. & Schielzeth, H. (2018). Condition-dependence and sexual ornamentation: effects of immune challenges on a highly sexually dimorphic grasshopper. Insect Science 25: 617-630. doi: 10.1111/1744-7917.12448External link
Valverde, J.P. & Schielzeth, H. (2015). What triggers colour change? Background colour and temperature effects on the development of an alpine grasshopper. BMC Evolutionary Biology 15: 168. doi: 10.1186/s12862-015-0419-9External link